Chapter: Transcription and Translation
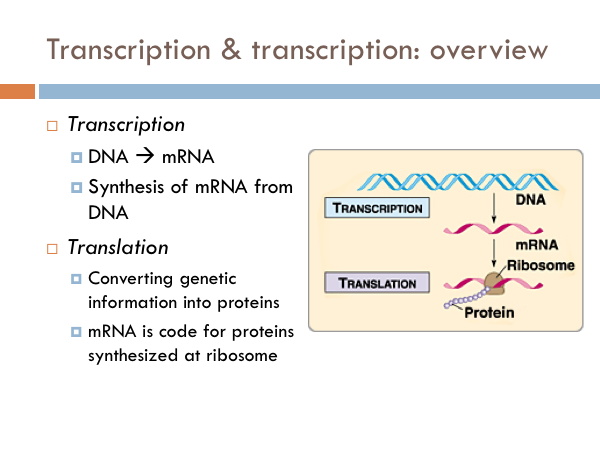
Transcription
In transcription, the strand of DNA that is used to synthesize mRNA is known as the template strand. Whereas, the non-template or coding strand matches the sequence of the RNA. However, it doesn’t match it exactly as RNA has uracil (U) instead of thymine (T). The nucleotides of RNA are known as ribonucleotides. These nucleotides bond to the template strand via hydrogen bonds after the DNA molecule opens up. And then those nucleotides are bonded together with a phosphodiester bond just like DNA is bonded.
RNA polymerase
RNA is an enzyme that synthesizes RNA from the template strand of DNA. And it happens a lot like DNA polymerase, except for the the fact that it does not require a primer before transcription begins Bacteria have a single RNA polymerase, whereas Eukaryotes have three different enzymes.
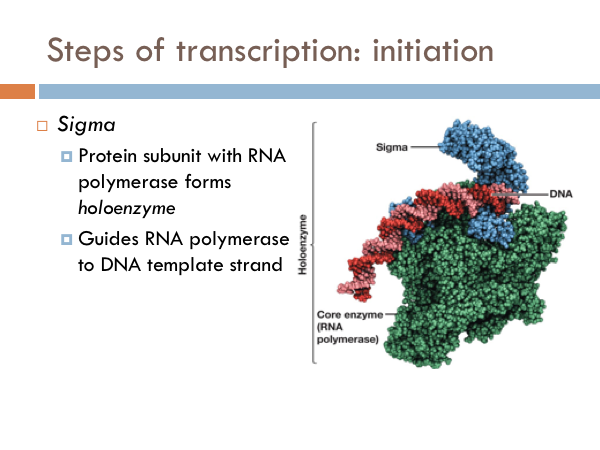
Initiation of Transcription
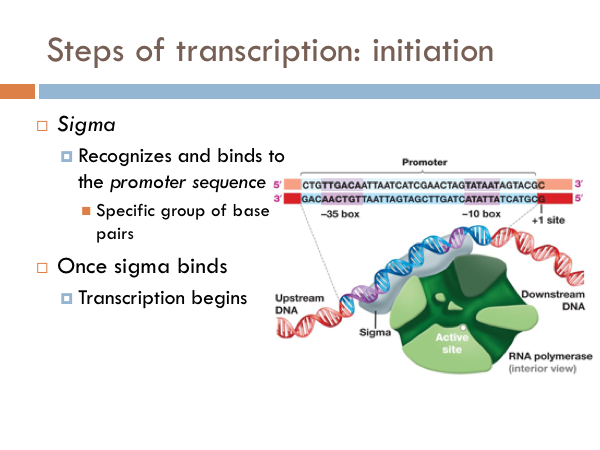
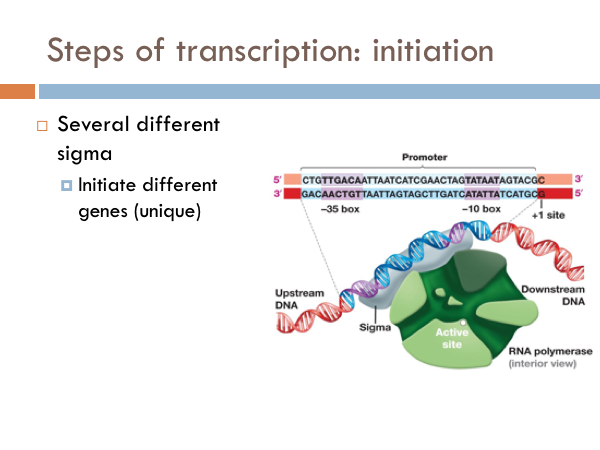
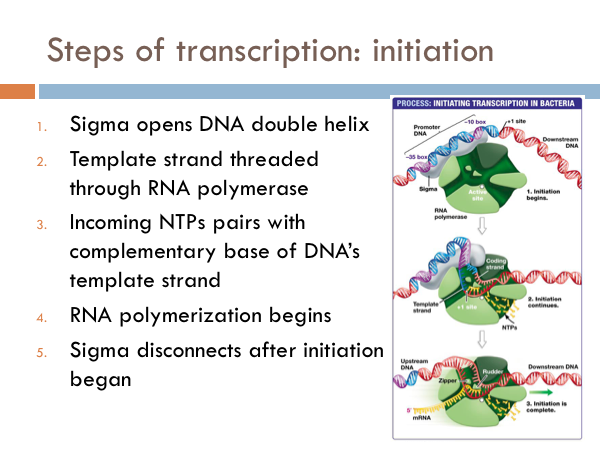
Transcription is initiated by the attachment of a protein known as a sigma. The sigma attaches to one strand of the DNA (the template strand) at a very specific location. In bacteria, several sigmas exists and each one initiates the transcription of a specific sequence of DNA (or gene). Once this sigma protein attaches to the DNA molecule, it serves to guide the RNA polymerase down the template strand. The sigma protein recognizes and binds to what is deemed the promoter sequence. The promoter sequence is a specific group of base pairs. Once the sigma binds to the DNA, transcription begins. There are several different sigmas. Each one is unique and initiates the synthesis of a specific gene, or in some cases several different genes. While there are several sigmas, each for different gene complexes, RNA Polymerase is the same molecule that connects to all the different sigmas. RNA Polymerase adds ribonucleotides to the template strand based on complementary base pairing, generating an mRNA.
The sigma protein first opens DNA’s double helix at the promoter section of the DNA strand. Then the template strand of the DNA is threaded through the RNA polymerase. Incoming RNA nucleotides come through a channel in the sigma protein and pairs with the complementary bases of the DNA’s template strand. At this point the RNA polymerase is functional and the begins to work. And once that happens the sigma disconnects from the DNA chain. This defines the beginning of the elongation phase of transcription.
Once the appropriate sigma is attached, RNA Polymerase attaches to the sigma protein. After successful attachment, the sigma guides the DNA into place inside of the RNA Polymerase. As the DNA is thread through the RNA Polymerase, hydrogen bonds are split between the the DNA molecule, by a zipper. Once DNA is inserted in to RNA Polymerase, ribonucleotides enter an entrance portal into the RNA Polymerase and match up with the D-nucleotides based on complementary base pairing. Similar to DNA base pairing, cytosine-containing deoxyribonucleotides (D-cytosine) pair with guanine containing ribonucleotides (R-guanine), D-guanine pairs with R-cytosine, and D-thymine pairs with R-adenine. Different from DNA base pairing, D-adenine pairs with R-uracil. Through another portal in the RNA Polymerase, emerges the developing mRNA. Once a few ribonucleotides are synthesized by RNA Polymerase, the sigma protein is removed. Once the sigma is removed, it can be reused to initiate transcription.
Elongation of Transcription
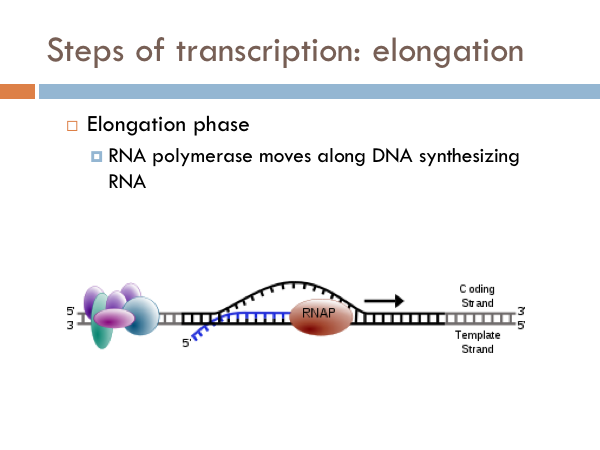
Elongation in transcription is fairly straight forward. The RNA polymerase zips along the open DNA molecule matching up complementary RNA base pairs from the template strand of the open DNA. After the sigma is removed, RNA Polymerase continues to unzip template and coding strands of the the DNA, and R-nucleotides are bonded via phosphodiester linkages using the code provided by the template strand of DNA. The incoming DNA enters into an intake portal and is unzipped by a zipper. As the DNA passes the zipper, the hydrogen bonds reattach between the coding and template strand and the DNA double helix leaves through an exit portal. Ribonucleotides enter in through another intake portal and are combined via complementary base pairing to the template strand of DNA. The R-nucleotides are bonded together via phosphodiester linkages. Ribonucleotides are continuously added to the 3’ end of the developing RNA strand. The 5’ end of the RNA strand leaves through another exit portal of the RNA Polymerase.
Termination of Transcription
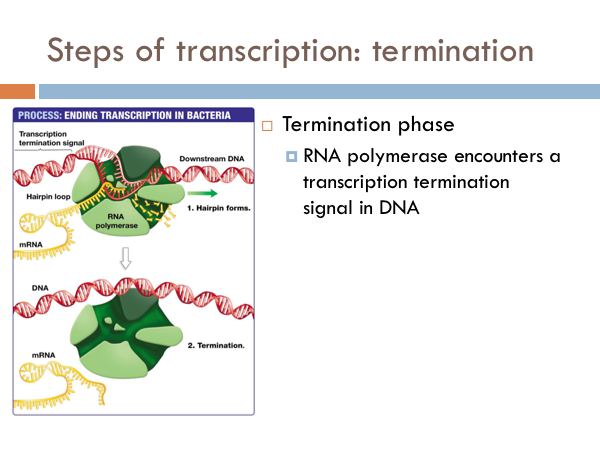
In bacteria, once RNA Polymerase transcribes a specific sequence of ribonucleotides from the DNA template strand, transcription ends (or terminates). When this sequence is synthesized, a section of the RNA bends back on itself forms a short double helix based on complementary base pairing. This forms a RNA hairpin. This hairpin forces the RNA to separate from the DNA and the RNA Polymerase detaches and the opened DNA reattaches based on complementary base pairing
Transcription in Eukaryotes
Fundamentally, transcription in eukaryotes is similar to transcription in prokaryotes with a few exceptions. In bacteria, RNA Polymerase can synthesize any RNA molecule. In eukaryotes, there are three different RNA Polymerases (I, II, and III). RNA Polymerase I is primarily responsible for the synthesis of ribosomal RNA (rRNA), the molecule that makes up ribosomes. Most eukaryotic RNA Polymerase are RNA Polymerase II. RNA Polymerase II is responsible for synthesizing mRNA, making it the only RNA Polymerase capable of transcribing protein-coding genes. RNA Polymerase III is responsible for synthesizing transfer RNA (tRNA). During translation, tRNAs read the messages from the mRNA and link a specific amino acid sequence generating proteins.
Where bacterial transcription is initiated by a sigma protein, RNA Polymerases in eukaryotes require a group of proteins known as basal transcription factors. Like sigma in prokaryotes, once the basal transcription factors attach to the DNA, its respective RNA Polymerase attaches and transcription begins. The elongation process is virtually identical in prokaryotes and eukaryotes. However, termination of transcription differs between prokaryotes and eukaryotes. In eukaryotes, a short sequence in the DNA signals the attachment of an enzyme downstream of active transcription. This enzyme cuts the emerging RNA, leaving the RNA Polymerase.
In eukaryotes, pre-RNA is made up of regions of mRNA that code for amino acids (known as exons) and regions of mRNA that don’t code for amino acids. Before the mRNA can be functional the introns must be removed in a process known as RNA splicing, or post-transcriptional modification.
Post-transcriptional modification of mRNA in eukaryotes
In bacteria, transcription from DNA to mRNA is a direct pathway. However in eukaryotes once mRNA is synthesized by RNA Polymerase II, the mRNA goes through further modification (Fig. 11). The product following transcription is known as a primary transcript (or pre-mRNA). Before mRNA travels outside the nucleus, the mRNA is shortened by cutting out specific sections of mRNA and reattaching the remaining sections back together. This process is known as RNA splicing and the resulting, modified mRNA is known as mature mRNA. Segments of the mRNA that are respliced back together are known as exons (because they exit the nucleus); while the segments of mRNA that are removed from the pre-mRNA are known as introns. The exons (which collectively make up the mature mRNA) leave the nucleus through a nuclear pore and travel to a ribosome in the cytosol and begin the process of translation.
RNA splicing is processed by hybrid protein-RNA complexes known as small nuclear ribonucleoproteins (or snRNPs). RNA splicing begins when a primary snRNP binds to a guanine R-nucleotide (G) adjacent to an uracil R-nucleotide (U) at the 5’ end of the pre-mRNA. This marks the exon-intron boundary. Another secondary snRNP reads from 5’ to 3’ down the mRNA and when it comes in contact with an adenine (A), and it attaches at that point. This point represents the intron-exon boundary. Once the primary and secondary snRNPs are attached other snRNPS attach to those, in a complex known as a spliceosome. Collectively the spliceosome breaks the G-U bond of the primary snRNP and the bond between the adenine (A) of the secondary snRNP and its adjacent R-nucleotide. Since U and A are complementary bases, the spliceosomes places them in close contact with each other, generating an intron loop. Nucleotides of the intron loop are disassembled into their monomers, ribonucleotides, and are recycled for future transcriptional events. Exons are spliced back together generating a mature mRNA.
Translation
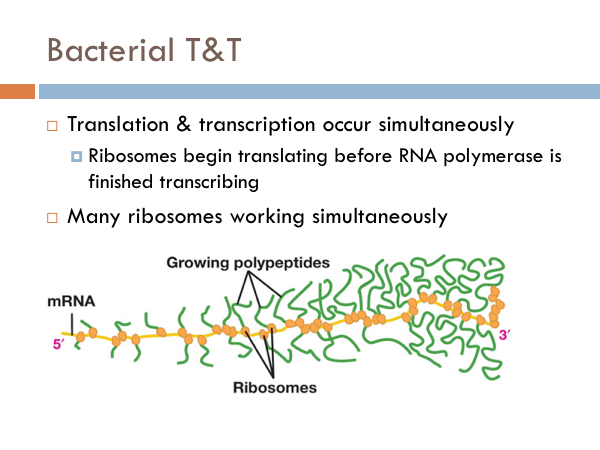
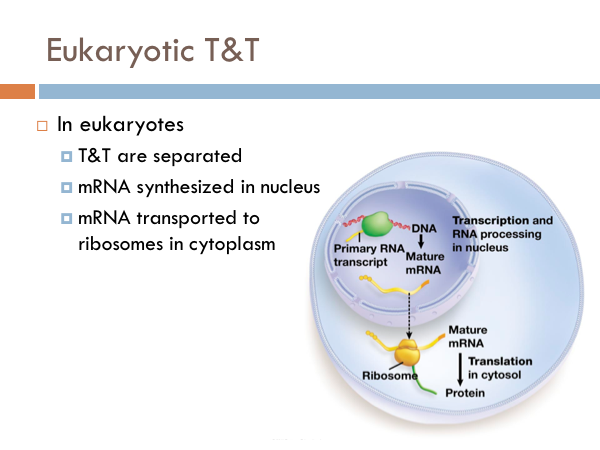
While transcription is the process of creating mRNA from DNA, translation is the process of converting the genetic information of mRNA into proteins. In bacteria, translation and transcription happen simultaneously. Ribosomes in proteins are floating right next to DNA. So in bacteria, the ribosomes begin the process of translation before the RNA polymerase terminates the transcription process. Another difference in bacteria is that many ribosomes are working simultaneously to synthesize proteins. In eukaryotic translation, transcription and translation are separated. mRNA are synthesized in the nucleus during transcription. The mRNA leaves the nucleus through the nuclear pore and travels to a ribosome in the cytoplasm, where the process of translation occurs. Most ribosomes are attached to the rough endoplasmic reticulum. However there are several ribosomes within the cytoplasm itself, as well.
Transfer RNA
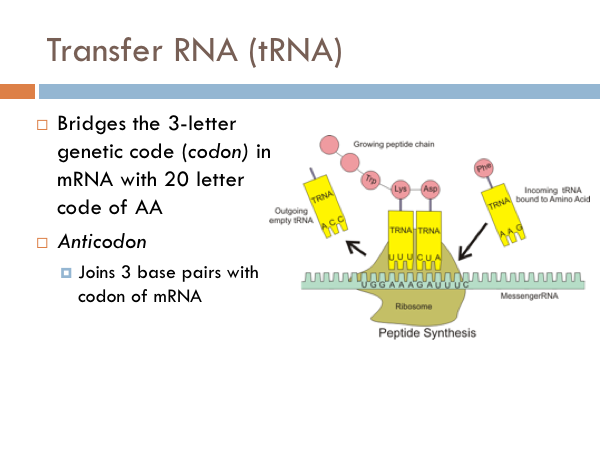
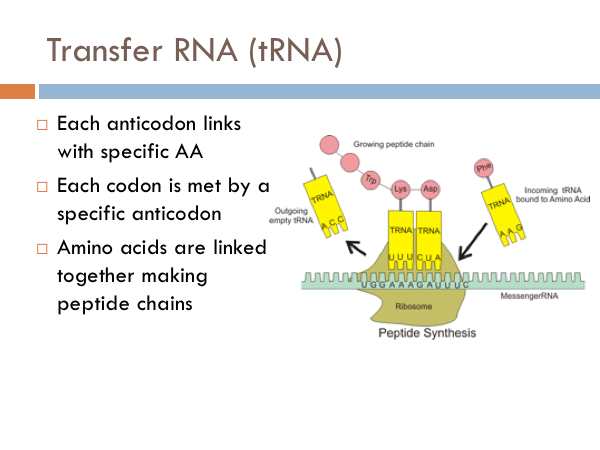
In addition to mRNA, another important RNA molecule is the transfer RNA, known as tRNA, tRNA is the molecule that bridges the genetic code with the a specific protein. Each transfer molecule is attached to a specific amino acid. And each amino acid has three base pairs attached to the opposite end of it.vAt the ribosome, the three base pairs of the tRNA join up with the complement of the three base pairs of the mRNA. So the three complimentary base pairs of the transfer RNA are known as an anticodon; whereas the triplet code of the messenger RNA is known as a codon. Each anticodon of tRNA links with a specific amino acid is combined to it complementary codon of mRNA at the ribosome. And then the amino acids are linked together by peptide bonds into a growing peptide chain.
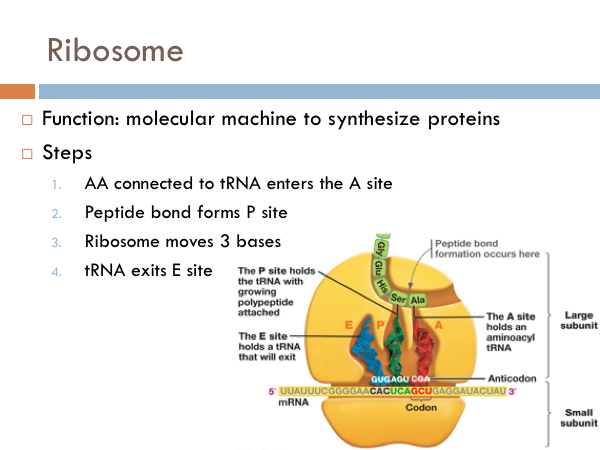
Ribosome
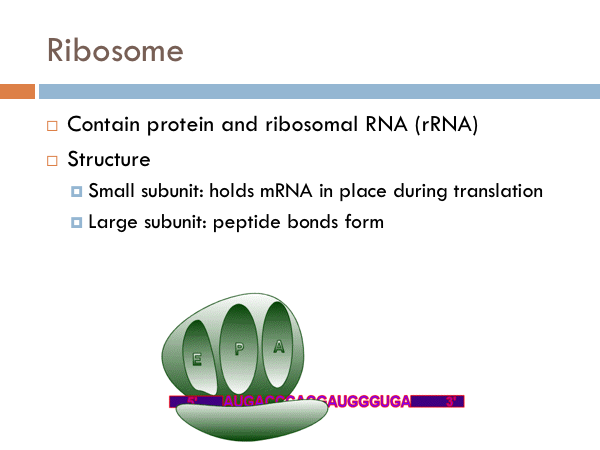
The ribosomal complex is made of another RNA (ribosomal RNA) and proteins. These make up two structure. The small subunit holds the mRNA in place during translation while the large subunit is where the peptide bonds form. And the large subunit has three distinct chambers: A, P and E. In general, the function of the ribosome is to synthesize proteins. First, the amino acid connected to a tRNA enters the ribosome at the A site. As another tRNA molecule comes into the A site, the other tRNA molecule moves over three base pairs and a peptide bond is formed between the two amino acids. As another tRNA molecule comes into the ribosomal complex, the other two tRNAs move 3 bases, and the oldest tRNA exits the ribosome at the E site. Just remember APE.
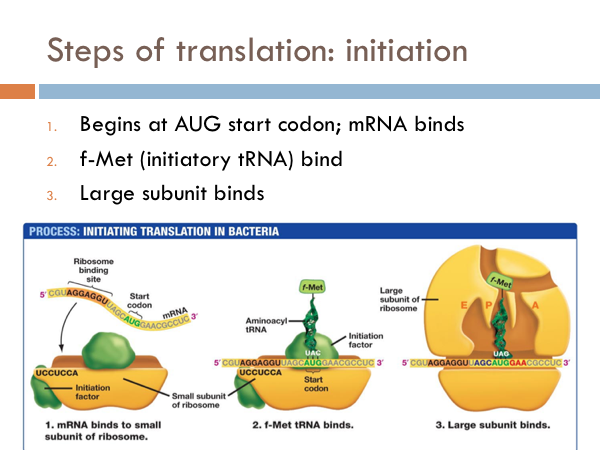
Initiation of Translation
Let’s take a closer look at the process translation. Translation is initiated by binding to a small subunit of the ribosome. The mRNA has a special section of it right before the start codon that the ribosome recognizes, known as the ribosome binding site. Translation begins with the start codon on the mRNA. Connected to the tRNA of the start codon is an amino acid, called f-Met. Once the f-Met tRNA binds to the small subunit of the ribosome, the large subunit of ribosome binds to the small unit, and translation is ready to begin.
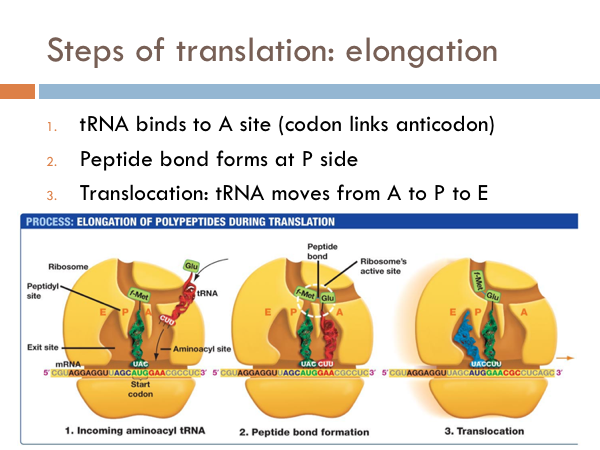
Elongation of Translation
Once the large subunit of the ribosome binds to the small subunit, the elongation process of translation begins. The first anticodon tRNA attached to an amino acid attaches to its complementary codon in the A site of the large subunit of the ribosome.
Second, a peptide bonds forms at the P site. Next the whole ribosome complex moves down three base pairs. The tRNA in the A site moves to the P site and the tRNA of the P site move to the E site, and the tRNA at the E site leaves the ribosomal complex. This process is known as translocation.
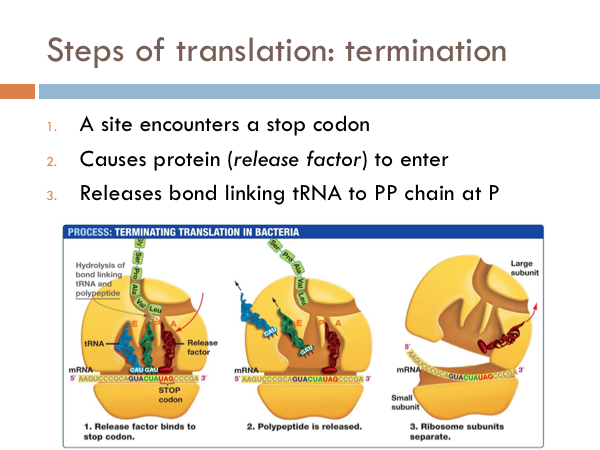
Termination of Translation
Translation is terminated by one of three stop codons. Once the ribosome encounters one of these stop codons, it causes a specific protein, known as a release factor, to enter the ribosome and it causes the release of the release of the polypeptide chain. Also at this time, the large ribosome separates from the small subunit.