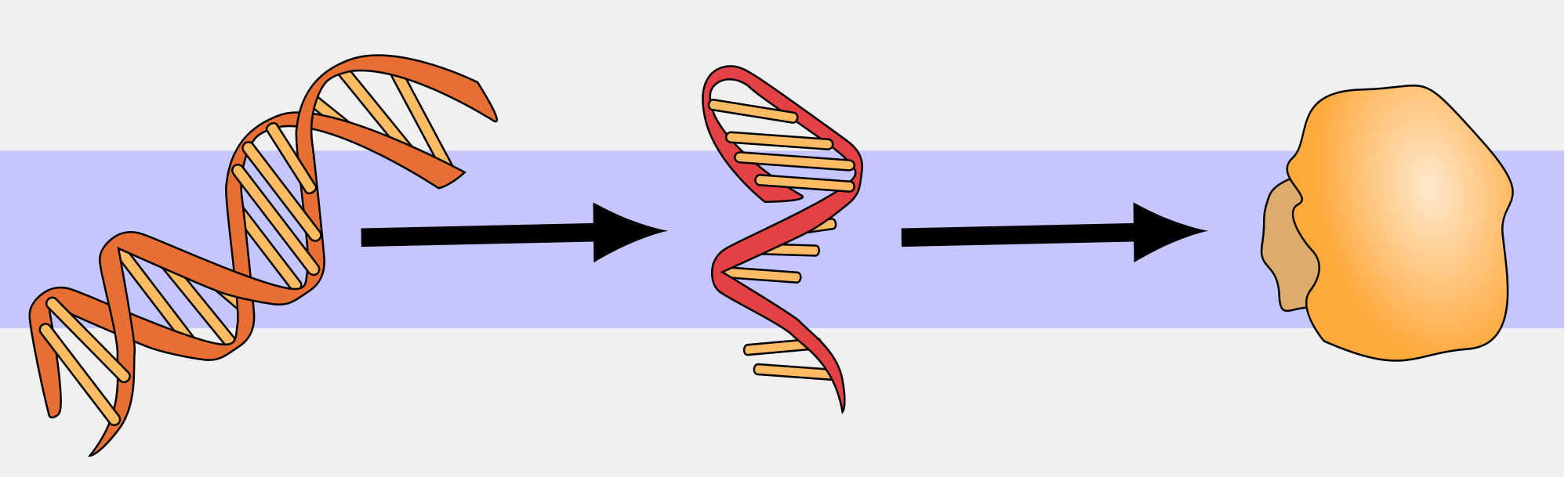
Transcription, RNA Processing and Translation
The Central Dogma of Molecular Biology
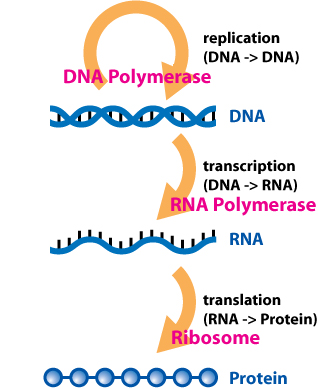
James Crick (cofounder of DNA’s secondary structure) proposed that DNA is an informational storage molecule capable of replicating itself. Further, he proposed that the information that was transmitted had to be “read” by a manufacturing body within the cell which put amino acids together in a specific sequence ultimately synthesizing a protein. This became known as the central dogma of molecular biology.
The central dogma of molecular biology suggests that DNA serves as a template for the direct synthesis of a messenger RNA (mRNA) molecule, in a process known as transcription. Secondly, mRNA is “read” at a ribosome by transfer RNAs (tRNAs) , which work together to assemble a specific chain of amino acids, which collectively assemble to generate a protein. This process is known as translation.
Exceptions to the Central Dogma of Molecular Biology
Reverse transcription
Reverse transcription is the transfer of information from RNA to DNA (the reverse of normal transcription). This is known to occur in the case of retroviruses, such as HIV, as well as in eukaryotes, in the case of retrotransposons and telomere synthesis. It is the process by which the genetic information from RNA will be assembled into new DNA.
RNA replication
RNA replication is the copying of one RNA to another. Many viruses replicate this way. The enzymes that copy RNA to new RNA, called RNA-dependent RNA polymerases, are also found in many eukaryotes where they are involved in RNA silencing. RNA editing, in which an RNA sequence is altered by a complex of proteins and a "guide RNA", could also be considered an RNA-to-RNA transfer.
Transcription in Prokaryotes
Transcription is the synthesis of mRNA from a DNA template. While DNA is double-stranded RNA is single stranded. Therefore, only one strand of DNA is copied, known as the template strand. The other strand of DNA is known as the non-template (or coding) strand.
Initiation of transcription: sigma
Like DNA Polymerase III, RNA is synthesized from 5’ to 3’. However unlike DNA Polymerase III, the molecule that synthesizes RNA (RNA Polymerase) does not require a RNA primer. Transcription is initiated by the attachment of a protein known as a sigma. The sigma attaches to one strand of the DNA (the template strand) at a very specific location. Since nucleotides can only be added from 5’to 3’, the connection of the sigma also determines the direction RNA Polymerase will travel. In bacteria, several sigmas exists and each one initiates the transcription of a specific sequence of DNA (or gene).
Once a sigma is attached to the template strand of the DNA, RNA Polymerase attaches to the sigma. While there are several sigmas, each for different gene complexes, RNA Polymerase is the same molecule that connects to all the different sigmas. RNA Polymerase adds ribonucleotides to the template strand based on complementary base pairing, generating an mRNA.
Initiation of transcription: RNA Polymerase
Once the appropriate sigma is attached, RNA Polymerase attaches to the sigma protein. After successful attachment, the sigma guides the DNA into place inside of the RNA Polymerase. As the DNA is thread through the RNA Polymerase, hydrogen bonds are split between the the DNA molecule, by a zipper.
Once DNA is inserted in to RNA Polymerase, ribonucleotides (R-nucleotides) enter an entrance portal into the RNA Polymerase and match up with the D-nucleotides based on complementary base pairing . Similar to DNA base pairing, cytosine-containing deoxyribonucleotides (D-cytosine) pair with guanine containing ribonucleotides (R-guanine), D-guanine pairs with R-cytosine, and D-thymine pairs with R-adenine. Different from DNA base pairing, D-adenine pairs with R-uracil. Through another portal in the RNA Polymerase, emerges the developing mRNA.
Initiation of transcription: removal of sigma
Once a few ribonucleotides are synthesized by RNA Polymerase, the sigma protein is removed. Once the sigma is removed, it can be reused to initiate transcription.
Elongation phase of transcription
After the sigma is removed, RNA Polymerase continues to unzip template and coding strands of the the DNA, and R-nucleotides are bonded via phosphodiester linkages using the code provided by the template strand of DNA. The incoming DNA enters into an intake portal and is unzipped by a zipper. As the DNA passes the zipper, the hydrogen bonds reattach between the coding and template strand and the DNA double helix leaves through an exit portal. R-nucleotides enter in through another intake portal and are combined via complementary base pairing to the template strand of DNA. The R-nucleotides are bonded together via phosphodiester linkages. R-nucleotides are continuously added to the 3’ end of the developing RNA strand. The 5’ end of the RNA strand leaves through another exit portal of the RNA Polymerase.
Termination of Transcription
In bacteria, once RNA Polymerase transcribes a specific sequence of R-nucleotides from the DNA template strand, transcription ends (or terminates). When this sequence is synthesized, a section of the RNA bends back on itself forms a short double helix based on complementary base pairing. This forms a RNA hairpin. This hairpin forces the RNA to separate from the DNA and the RNA Polymerase detaches and the opened DNA reattaches based on complementary base pairing.
Transcription in Eukaryotes
Fundamentally, transcription in eukaryotes is similar to transcription in prokaryotes with a few exceptions. In bacteria, RNA Polymerase can synthesize any RNA molecule. In eukaryotes, there are three different RNA Polymerases (I, II, and III). RNA Polymerase I is primarily responsible for the synthesis of ribosomal RNA (rRNA), the molecule that makes up ribosomes. Most eukaryotic RNA Polymerase are RNA Polymerase II. RNA Polymerase II is responsible for synthesizing mRNA, making it the only RNA Polymerase capable of transcribing protein-coding genes. RNA Polymerase III is responsible for synthesizing transfer RNA (tRNA). During translation, tRNAs read the messages from the mRNA and link a specific amino acid sequence generating proteins.
Where bacterial transcription is initiated by a sigma protein, RNA Polymerases in eukaryotes require a group of proteins known as basal transcription factors. Like sigma in prokaryotes, once the basal transcription factors attach to the DNA, its respective RNA Polymerase attaches and transcription begins. The elongation process is virtually identical in prokaryotes and eukaryotes. However, termination of transcription differs between prokaryotes and eukaryotes. In eukaryotes, a short sequence in the DNA signals the attachment of an enzyme downstream of active transcription. This enzyme cuts the emerging RNA, leaving the RNA Polymerase.
Post- transcriptional modification of mRNA in eukaryotes
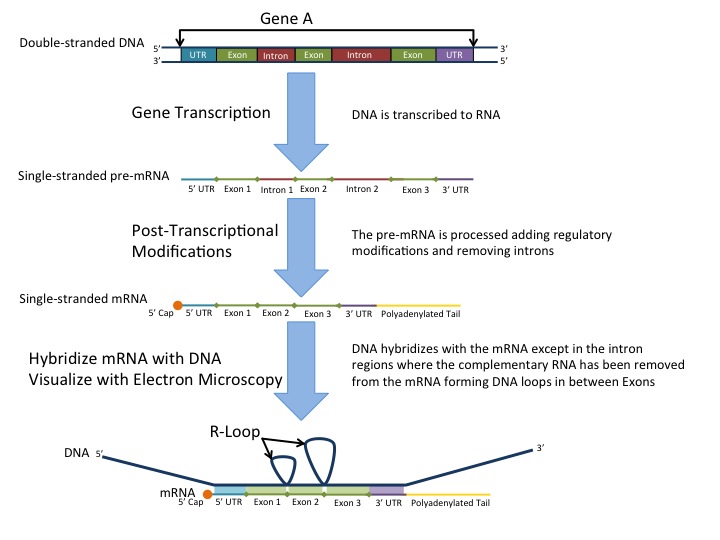
In bacteria, transcription from DNA to mRNA is a direct pathway. However in eukaryotes once mRNA is synthesized by RNA Polymerase II, the mRNA goes through further modification (Fig. 11). The product following transcription is known as a primary transcript (or pre-mRNA). Before mRNA travels outside the nucleus, the mRNA is shortened by cutting out specific sections of mRNA and reattaching the remaining sections back together. This process is known as RNA splicing and the resulting, modified mRNA is known as mature mRNA. Segments of the mRNA that are respliced back together are known as exons (because they exit the nucleus); while the segments of mRNA that are removed from the pre-mRNA are known as introns. The exons (which collectively make up the mature mRNA) leave the nucleus through a nuclear pore and travel to a ribosome in the cytosol and begin the process of translation.
RNA splicing is processed by hybrid protein-RNA complexes known as small nuclear ribonucleoproteins (or snRNPs). RNA splicing begins when a primary snRNP binds to a guanine R-nucleotide (G) adjacent to an uracil R-nucleotide (U) at the 5’ end of the pre-mRNA. This marks the exon-intron boundary. Another secondary snRNP reads from 5’à3’ down the mRNA and when it comes in contact with an adenine (A), and it attaches at that point. This point represents the intron-exon boundary. Once the primary and secondary snRNPs are attached other snRNPS attach to those, in a complex known as a spliceosome. Collectively the spliceosome breaks the G-U bond of the primary snRNP and the bond between the adenine (A) of the secondary snRNP and its adjacent R-nucleotide. Since U and A are complementary bases, the spliceosomes places them in close contact with each other, generating an intron loop. Nucleotides of the intron loop are disassembled into their monomers, R-nucleotides, and are recycled for future transcriptional events. Exons are spliced back together generating a mature mRNA.
The Genetic Code
Triplet code hypothesis
Once it was determined that DNA’s main function was to code for proteins, the next question was how. The language of DNA is based on four nucleotides (A, T, C, and G), while proteins are composed of 20 amino acids. Thus, it is mathematically impossible for one nucleotide to code for one amino acid, as there are more than four amino acids.
A + T + C + G = 41 = 4 < 20
Therefore, amino acids must be coded by some combination of D-nucleotides. What if DNA was coded by a combination of two neighboring nucleotides?
AA + AT + AC +AG + TA + TT + TC + TG + CA + CT + CC + CG + GA +GT + GC + GG = 42 = 16 < 20
This mechanism also results in fewer than 20 amino acids. Therefore, it was hypothesized that the simplest possible code would have to be based on a triplet combination of neighboring D-nucleotides.
The code is redundant
As you see from your calculation, there are far more possibilities of amino acids provided by a triplet code, than the number of amino acids (20) we see in nature. Therefore, it is said that the code is redundant, meaning that amino acids can be coded by more than one triplet code. For example, the triplet codes of CCU and CCC of mRNA code for the same amino acid, proline. In fact, all amino acids are coded by more than one triplet code except for methionine and tryptophan.
The code is unambiguous
Further investigations indicated that a specific triplet code always coded for the same amino acid. In other words, the code is unambiguous. For example, the triplet code of AUG in mRNA always codes for methionine.
The code is universal
Amazingly, the code works exactly the same for all living organisms, from bacteria to plants and animals! While there are very few exceptions to this, the consistency of the code across widely variable organisms hints that we all stem from a single common ancestor.
The Genetic Code. The triplet code mRNA directly codes for the assembly of amino acids that make up a protein. To identify the amino acid coded by the mRNA sequence, locate the mRNA triplet code (codon), the grey box to its right represents the corresponding amino acid. For example, CCC indicates the amino acid Proline (Pro).
Translation
Translation in prokaryotes
Translation occurs at ribosomes in all cells. Since prokaryotic DNA is not bound by a nucleus, translation in prokaryotes occurs before transcription is complete. Transcription and translation occur simultaneously. This has the advantage of being much faster than in eukaryotes.
Translation in eukaryotes
In eukaryotes, transcription and modification of mRNA happens exclusively in the nucleus. After mRNA processing, the mature mRNA travels out of the nucleus through a nuclear pore. In the cytosol, the liquid body of the cell outside the nucleus, the mature mRNA attaches to a ribosome and goes through translation. It is thought that this separation of transcription and translation provides a greater control over gene regulation, specifically by the removal of introns from the pre-mRNA. It is also hypothesized that eukaryotic DNA is less susceptible to mutations than prokaryotes, due to the physical barrier of the nuclear envelope between the DNA and the cytosol.
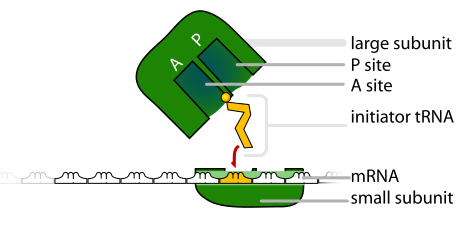
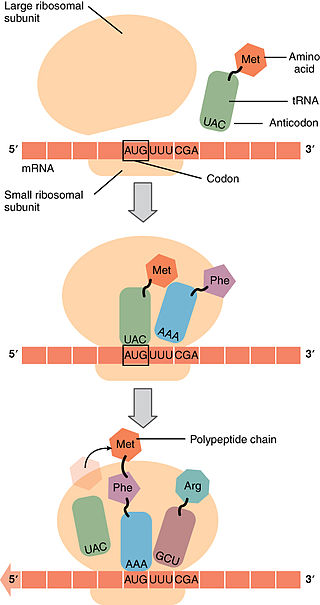
Initiation of translation
Translation begins when an mRNA connects to the small subunit of a ribosome. Ribosomes are made up of proteins and another type of RNA, ribosomal RNA (or rRNA). Initiation of translation begins when rRNA binds to a specific sequence of the mRNA, known as the ribosome binding site. This connection is based on complementary base pairing of adjacent R-nucleotides of rRNA and and mRNA, which is guided into place by special proteins known as initiation factors. One of the initiation factors also serves as a docking station for the first tRNA to connect to the start codon of mRNA, which is AUG for the synthesis of all proteins. tRNAs have a complementary triplet code that connects to the codon of the mRNA, known as an anticodon. The anticodon of the initial tRNA is UAC. Attached to the initial tRNA is the amino acid, Methionine (Met). Once the anticodon (UAC) of the initial methionine-containing tRNA is successfully paired to the complementary start codon of the mRNA (AUG), the large subunit of the ribosome attaches to the mRNA. The initial tRNA (with the anticodon, UAC) acts like a key, locking the small and large subunits together with the mRNA sandwiched between. Once both subunits are attached, the initiation factors are removed. After the initial tRNA signals the attachment of the large subunit of the ribosome, all subsequent tRNAs enter the large subunit through its A site. The next tRNA enters the A site due to complementary base pairing of the codon of the mRNA and the anticodon of the tRNA. Once the codon-anticodon pairing is successful, the new tRNA in the A site is positioned such that the amino acid it is carrying is adjacent to the amino acid already present in the P site. This proximity encourages a peptide bond to form between the two adjacent amino acids.
Elongation of translation
Once the peptide bond forms between the first two amino acids, the mRNA moves three base pairs from the A site towards the E site. The initial tRNA moves from the P site to the E site and the second tRNA moves from the A site to the P site. A new tRNA enters into the A site due to complementary base pairing based on the codon. Another peptide bonds forms between the newly adjacent amino acids connected to the tRNAs in the A and P site.
Once the second peptide bond forms, the mRNA again moves downstream (5’ to 3’) on the ribosome. The tRNAs move from A site to P site to E site. In the next move the tRNA in the E site is ejected from the ribosome. The elongation process repeats this process thousands of times until the mRNA reaches a termination signal.
Termination of translation
The termination signal is known as a stop codon. Unlike all the other codons, the tRNA complementary to the stop codon does not have an associated amino acid connected to it. This tRNA is known as a release factor and it breaks the bond linking the tRNA to the amino acid in the P site, releasing the polypeptide chain. Once the protein is released from the ribosome, the tRNAs, mRNA, large subunit and small subunit of the ribosome all disassemble. The protein is fully functional (unless further modification is required), and the mRNA can again enter translation producing more proteins.