Module Instructions: DNA Replication (Lab)
Pre-Lab: DNA Replication
Watch the lecture. Read the chapter and complete the study guide in preparation for Exam 1. Exam 1 is next week. If you have any questions about the study guide, please reach out to me through Canvas Inbox. I am here to help you understand the materials and reach your goals.
Lab: DNA Replication
This lab can be a challenge. Below are points for the most commonly made mistakes in the lab.
Just as a reminder, you are responsible for completing all of the items in the rounded boxes with the check marks: ☑. In Figure 1, the replication forks are where the active site of DNA Replication is occurring. There are two replication forks per replication bubble.
For Problem 1, you are to use the same methodology to straighten a spring that topoisomerase uses in DNA Replication. Think of a slinky. You can’t just uncoil it. It would supercoil. What topoisomerase does is it cuts the DNA as Helicase is opening the DNA strands. Once this cut is made, the tension of the supercoiling effect is relieved and topoisomerase reattaches (welds, if you will) the DNA strands back together. This process prevents the DNA from supercoiling.
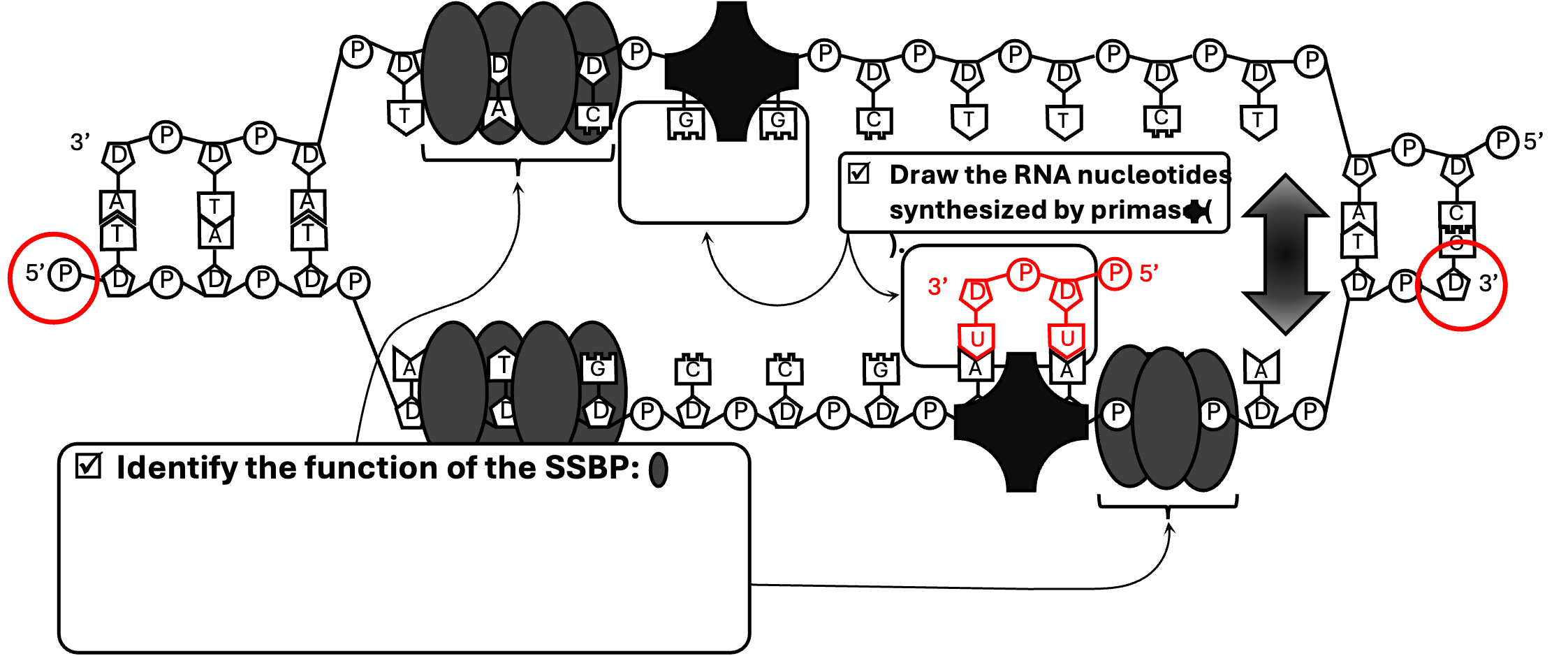
In Figure 6, you will draw the new ribonucleotides synthesized by RNA Primer. Remember a ribonucleotide has three components: a phosphate, a ribose (sugar) and one of the four nitrogenous bases: A, U, C or G. The new strand that is created will be antiparallel to the existing strand. Below is a demonstration of how to complete this for the bottom strand (which you will soon discover is the lagging strand). In this case the bottom (original) strand has a phosphate on the far left side (the 5’ end) and a ribose on the far right side (the 3’ end). When the new ribonucleotides bind to the existing deoxyribonucleotides based on complementary base pairing, the new ribonucleotides are flipped (antiparallel). In this case the phosphate of the ribonucleotides are facing the opposite direction as the existing DNA strand. Repeat this strategy to complete the ribonucleotides for the top strand.
In Figure 7, the leading strand is the one that is synthesized continuously; whereas the lagging strand is synthesized discontinuously.
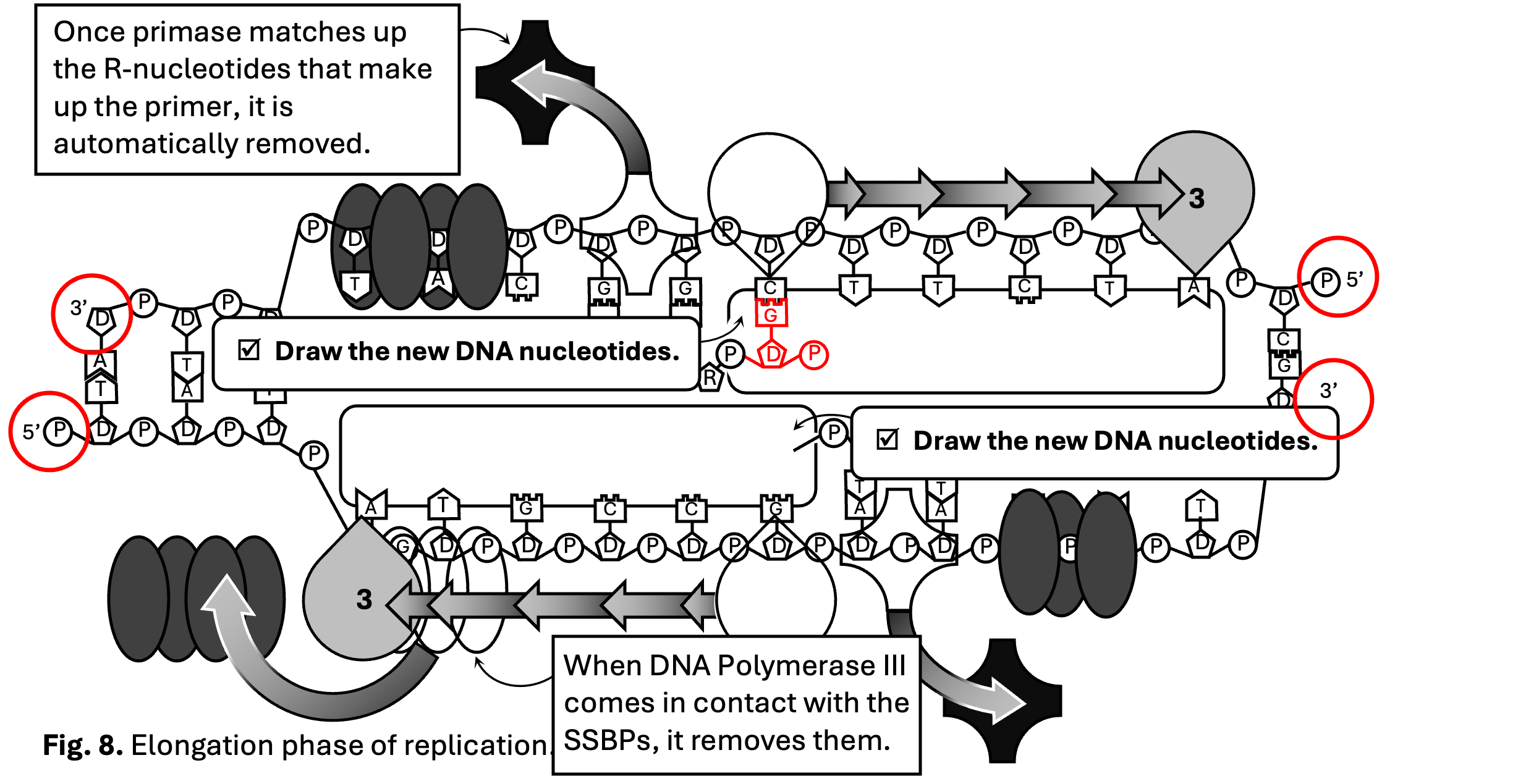
In Figure 8, use the same strategy as you used for the ribonucleotides in Figure 6 to input the new deoxyribonucleotides. Remember a deoxyribonucleotide also has three components: a phosphate, a deoxyribose (sugar), and a nitrogenous base (A, T, C or G). Also remember the phosphate must face the opposite direction as the existing (or parental) strand in order to make the two eventual strands antiparallel. In the top strand, the phosphate is on the right side of the image. Therefore, the newly synthesized deoxyribonucleotides will face the opposite direction, to the left. Below, you will see the first deoxyribonucleotide of the top strand (which you will soon see is the leading strand).
In Figure 9, there is some confusion as to which is the leading and which is the lagging nucleotides. If you look at Figure 10 and 11, you will notice the top strand continuously synthesizes; this is the leading strand. Wheras the bottom strand is synthesized in fragments, the lagging strand.
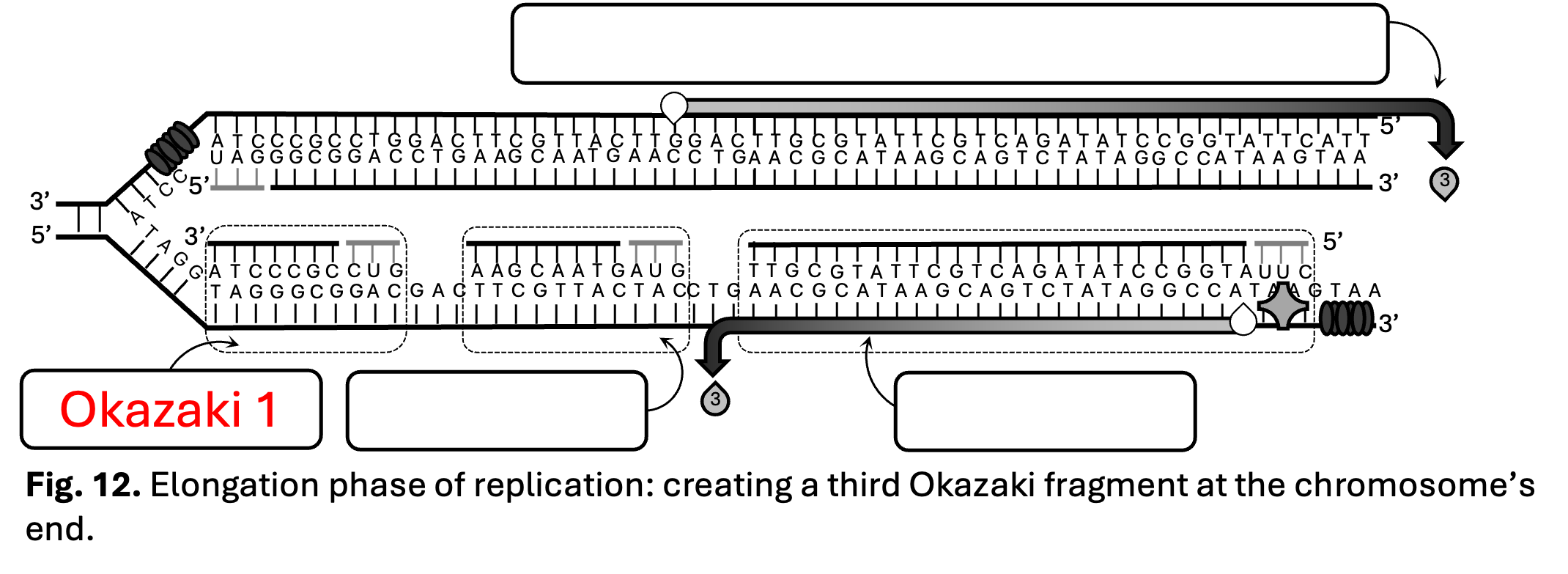
In Figure 12, you are to identify the order of the Okazaki fragments. If you look back at Figures 9 through 11, you will notice that the left most Okazaki fragment was synthesized first. Therefore that would be Okazaki fragment 1.
In Figure 14, no need to overthink this one. Ligase connects the Okazaki fragments. It should look something like this.
For Problem 8, the reason there are unsynthesized nucleotides in the lagging strand after telomerase extends it is the same reason there are unsynthesized nucleotides before the telomerase extends it. In other words, your answer for Problem 8 will be exactly the same as Problem 5.
Post-Lab: DNA Replication
Once you finish your Pre-Lab and Lab, complete your Post-Lab by the deadline in the class calendar.