DNA replication is the process of producing two identical replicas from one original DNA molecule. This biological process occurs in all living organisms and is the basis for biological inheritance. DNA is made up of two strands and each strand of the original DNA molecule serves as template for the production of the complementary strand, a process referred to as semiconservative replication. Cellular proofreading and error-checking mechanisms ensure near perfect fidelity for DNA replication.
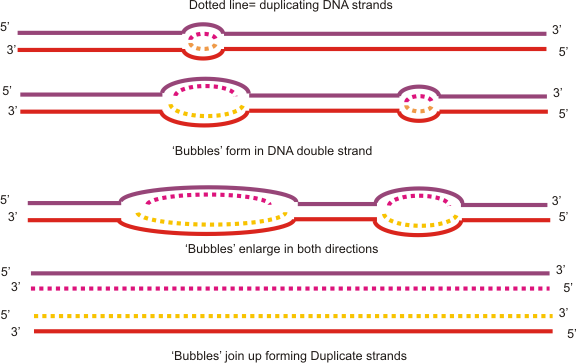
Replication bubbles
DNA replication is the process in which a DNA molecule makes a copy of itself. DNA replication differs in minor ways between prokaryotes and eukaryotes. Prokaryotes are single-celled organisms that lack a nucleus (i.e. bacteria). All prokaryotes have circular DNA which replicates from a single origin, resulting in a single replication bubble. The replication bubble grows in two directions from its origin. There are two replication forks in a replication bubble, which is where the parental DNA double helix is split. Eukaryotes are organisms that have a true nucleus, ncluding unicellular and multicellular organisms. In the nucleus, eukaryotic DNA is linear and it replicates from several origins, resulting in several replication bubbles.
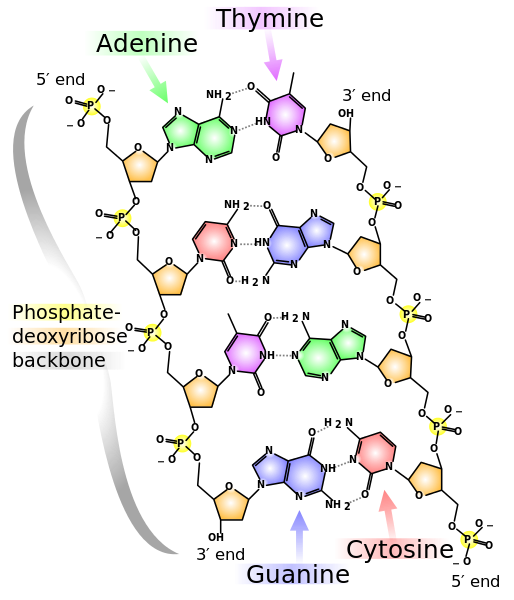
DNA prior to replication
Prior to replication, DNA is a double stranded helix with complementary base pairing. In order to achieve complementary base pairing, the two strands of DNA are antiparallel, or line up in opposite directions. On one strand the phosphate group is on one end of the DNA backbone while the deoxyribose is at the opposite end. In the second strand, phosphate is on the end in which the deoxyribose of the first strand is located, while the deoxyribose end of the second strand is adjacent to the first strand’s phosphate end. The phosphate end of a DNA strand is called the 5’ end of the DNA backbone, while the deoxyribose end is the 3’ end.
Initiation of replication
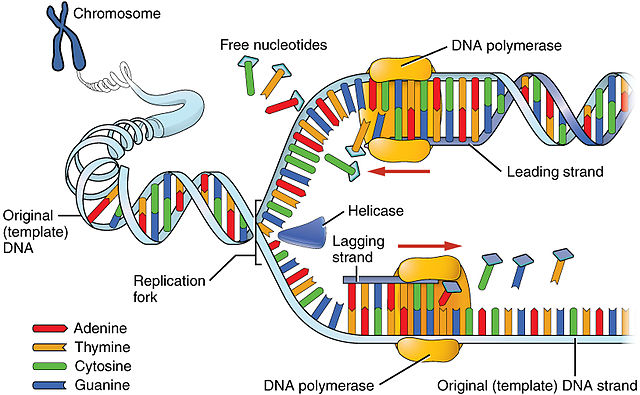
The replication bubble begins with the enzyme, helicase, breaking the hydrogen bonds between nitrogenous bases of the two parental DNA strands (Fig. 5). In order to prevent the hydrogen bonds from reforming, single-strand DNA-binding proteins (SSBPs) attach to the separated DNA strands (Fig. 6). The breaking of the parental DNA’s hydrogen bonds causes the double helix to relax, creating tension further down the molecule. Another enzyme, topoisomerase, cuts the covalent bonds (phosphodiester linkages) of the double helix further downstream of the replication bubble and immediately reattaches them, relieving the tension.
Once the parental DNA strands are opened and stabilized, the enzyme primase adds a few ribonucleotides (R-nucleotides) to the template strands of the parental DNA (Fig. 6). These RNA nucleotides are primers allowing DNA Polymerase III to attach. DNA Polymerase III is the main workhorse of DNA replication, matching up D-nucleotides to the opened parental DNA strand. The RNA primer allows DNA Polymerase III to attach and provides a hydroxyl (OH) group so the first D-nucleotide can be matched up with the parental strand.
Elongation stage of replication: leading strand vs. lagging strand
Once DNA Polymerase III is in place, primase is removed and new D-nucleotides can only be added to the 3’ carbon of the deoxyribose in the parental strand. From the perspective of the parental strand new DNA grows from the 5’ end of the nucleic backbone towards the 3’ end, from 5’ à 3’. As helicase opens the DNA, DNA Polymerase III continually adds D-nucleotides in one direction. This strand is known as the leading strand. As helicase opens the double helix, the leading strand is continuously replicated by DNA Polymerase III.
Elongation stage of replication: leading strand vs. lagging strand (con’t)
On the opposite strand a problem arises due to the antiparallel nature of DNA and the fact that DNA Polymerase III can only add base pairs in the 3’ direction. SSBPs must be added to prevent the parental DNA strands from reattaching, primase has to attach another RNA primer and DNA Polymerase III has to add D-nucleotides in the reverse direction as the helicase is traveling. This disrupts the addition of D-nucleotides by DNA polymerase III, causing the initiation and elongation steps of DNA replication to repeat multiple times. This discontinuously generated new DNA strand is the lagging strand.
Completion stage of replication: DNA Polymerase I
Once DNA Polymerase III pairs the majority of complementary D-nucleotides on the developing leading and lagging strand, the RNA primers are still present. Another DNA Polymerase enzyme, DNA Polymerase I reads both the leading and lagging strands from 5’ to 3’ of the parental strands replacing the R-nucleotides with D-nucleotides. While the leading strand is synthesized continuously, there is still an RNA primer at the origin of the replication bubble. It is also replaced by DNA Polymerase I.
Completion stage of replication: Ligase
After DNA Polymerase I replaces the RNA primers, there is no phosphodiester linkage between Okazaki fragments. Another enzyme, ligase, travels down the lagging strand creating a phosphodiester linkage connecting the Okazaki fragments.
Completion stage of replication: Telomerase
In prokaryotes, once ligase is finished connecting the Okazaki fragments, the DNA has been successfully replicated. This is due to the circular shape of their DNA. However, eukaryotic DNA is linear. In the lagging strand this poses a problem. At the end of the chromosomes, known as the telomeres, there is nowhere for the DNA Polymerase III to connect due to the absence of an RNA primer. If these D-nucleotides at the telomeres were left unsynthesized, the DNA strands would eventually become shorter with every replication. It is thought that aging is a product of malfunctioning telomerase.
Telomerase is an enzyme that attaches to the unsynthesized end of the lagging strand and catalyzes the synthesis of DNA from its own RNA template, akin to reverse engineering. At one end of the telomerase a few R-nucleotides combine with the end of the unreplicated strand. Telomerase then adds D-nucleotides to the end of the lagging strand.
The opposite ends of telomerase are identical. The opposite side of the telomerase (if you read left to right) ends with the same R-nucleotides (AUU). Telomerase uses this phenomenon to detach once the lagging strand has been extended and then reattach back to the end of the extended lagging strand. The reattached telomerase replicates another exact copy to the lagging strand. This process happens several times and the telomerase eventually is removed.
Once telomerase lengthens the parental lagging strand, primase attaches synthesizing an RNA primer, which allows DNA Polymerase III to replicate the remaining D-nucleotides on the unreplicated strand. Ligase fuses the last remaining segment by creating the final phosphodiester linkage. This results in two exact copies of the original DNA with slightly longer telomeres.
Repairing mistakes
DNA Polymerase III is extremely accurate. Another DNA Polymerase acts as a proof reader, DNA Polymerase II. Once replication is complete, DNA Polymerase II travels down the entire link of the parental DNA strands looking for mismatched base pairs. Once it detects a mismatch, DNA Polymerase II removes the D-nucleotide from the new DNA strand and replaces it with the D-nucleotide complementary to the parental strand.
However, even this proofreading step is not completely accurate, missing mismatched base pair occurs approximately one out every billion times. These mistakes lead to nucleotide-level mutations. Such mutations are the only way in which new alleles are generated, and are usually neutral or deleterious relative to fitness. Occasionally these mutations will create beneficial phenotypes allowing these organisms higher probability of surviving and reproducing. Such mutations tend to amplify in future generations due to natural selection.