Chapter: DNA Synthesis
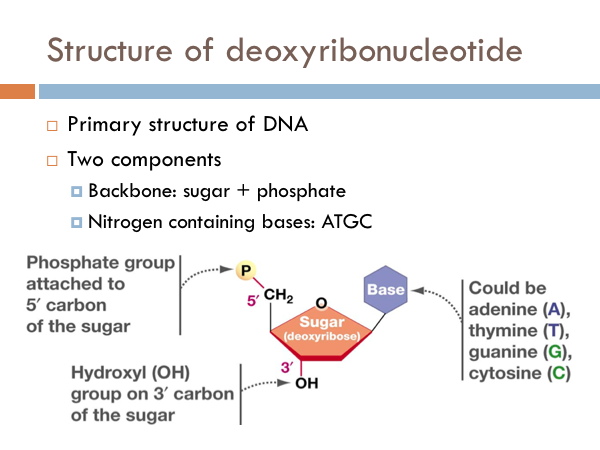
The simplest component of DNA is a nucleotide. More precisely, it is a deoxyribonucleotide. It has a backbone made of a phosphate group attached to the 5’ carbon of a sugar. These form the backbone of the DNA, or the sides of the ladder. The code of DNA is determined by the nitrogenous bases. And these could be made of adenine, thymine, guanine, or cytosi
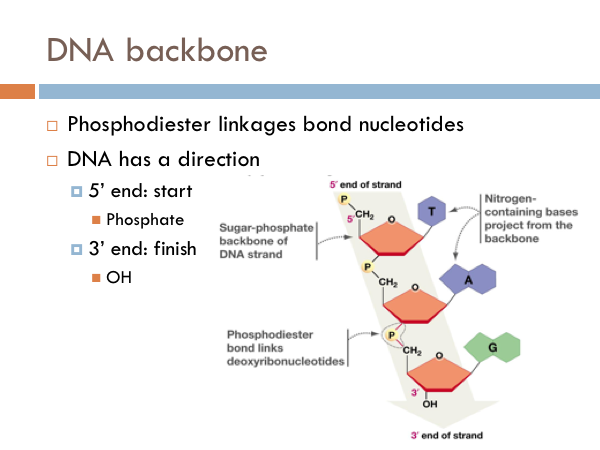
DNA Backbone
Each nucleotide is linked to another by what is known as a phosphodiester linkage. And DNA has a direction. DNA is read from a 5 prime to 3 prime direction. What this means is that DNA is read much like we would read a book. It starts at the 5 prime end of the strand. That is the strand that begins with the phosphate group. And then DNA is read toward the 3 prime direction, or towards the sugar end of the DNA.
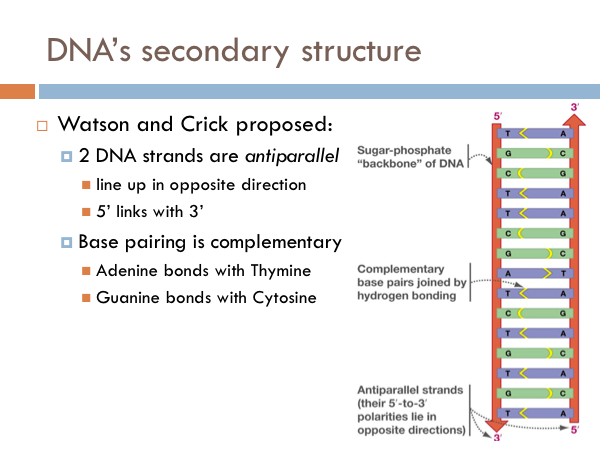
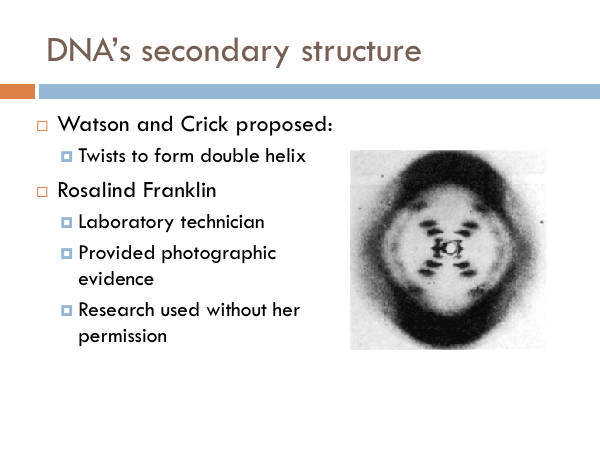
DNA’s Secondary Structure
Not much was known about DNA, until the 1950s. We knew that DNA was the code of life. But we had no idea how it worked. All we really knew was that it was made of sugar, phosphate, and the four nitrogenous bases. Much of the foundational work on DNA was done by James Watson and Francis Crick. In 1953, Watson and Crick identified the structure of DNA. Throughout their collaboration, Watson and Crick proposed three characteristics.
It was known that the two backbones of the DNA molecule line up side by side. However, they could theoretically line up two different ways. They could line up in the same direction, that is both sides of the ladder would be 5 prime to 3 prime in the same direction. Watson and Crick, however, proposed that the 2 strands of DNA are antiparallel. This doesn’t mean that they were perpendicular. Rather, it means that the line up side by side, but the direction of the DNA backbone goes two different directions. And if this were to be true, Watson and Crick proposed that the pairing of the nitrogenous bases was complementary. That is, adenine bonds with thymine and guanine bonds with cytosine. If DNA were parallel rather than antiparallel, it is theoretically possible that like nitrogenous bases paired with each other. That would be an A with an A, a C with a C, and so on.
Shape of DNA
Watson and Crick later shared a Nobel Prize. Their discovery depended heavily on the work of a woman, chemist Rosalind Franklin, whose research was used without her knowledge or permission. Even worse, she wasn’t given credit for her work. She was not authored on their landmark publication identifying the structure of DNA. It was Franklin's photograph of the DNA molecule that sparked a scientific revolution spearheaded by Watson and Crick. In regards to first seeing this photoWatson said, "My jaw fell open and my pulse began to race." The photo showed, for the first time, the essential structure of DNA -- the double-helix shape, which also indicated its method of replication. It was Franklin's photographic skills that made the discovery possible. She did not know the other men were using her research upon which to base the article that appeared in the journal Nature. She wouldn't share in the Nobel Prize either. However this is not because Franklin was overlooked, but because she was dead. The award is not given out posthumously. Franklin was diagnosed with ovarian cancer in 1956 at age 37, and died two years later (likely because of her exposure to radiation during her laboratory work.
DNA as templates
As scandalous as the situation was with Rosalind Franklin was, James Watson and Francis Crick were truly revolutionary scientists. Watson and Crick proposed that the two strands of DNA served as templates (or patterns) for the production of new strands of DNA. In this way, DNA could be copied over and over and over again. Life requires this. They argued that these strands are copied according to this complementary base pairing. In other words, a DNA molecule opens up and the nitrogenous base pairs match up with their complement (As with Ts, and Cs with Gs).
Hypotheses of DNA Replication
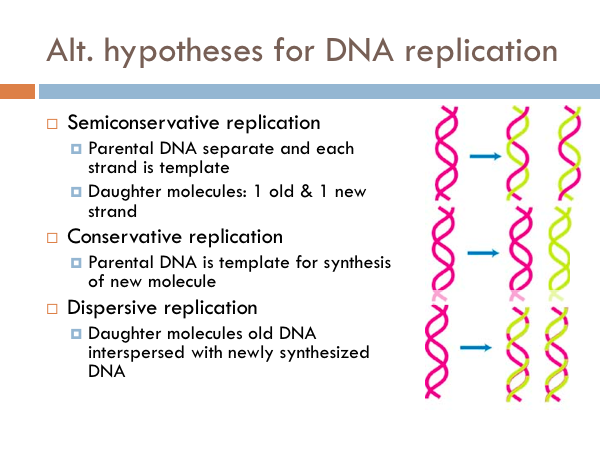
Semiconservative replication
Once the structure of DNA was established, the next major feat was to understand how it was replicated. And there are three possible ways DNA could replicate. These are known as the alternate hypotheses for DNA replication. The hypothesis of semiconservative replication predicts that the parental DNA separates and each strand serves as a template, in which a new copy is made from each of the old strands. So in the picture the pink strands represent the parental DNA strands; whereas the yellow strand represents the copied chromosomes (the daughter chromosomes). The pink strands separate, a copy is made and then each new double strand of DNA has a parental strand and a daughter strand.
Conservative replication
In conservative replication, the parental DNA is used as a template for the synthesis of new molecule. In other words, the both parental DNA strands are copied, and we would end up with two strands of DNA, one was the original parental DNA and the other is a complete copy of the parental double strand of DNA.
Dispersal replication
The other possibility for DNA replication is known as dispersive replication. In this model, the new DNA molecules are a combination of segments of parental DNA and daughter DNA. … So how is DNA replicated. We know based on the results of an ingenious experiment.
Meselson-Stahl Experiment
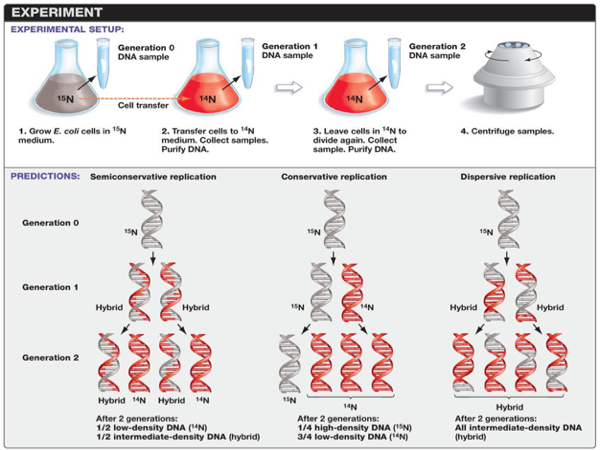
In 1958, Matthew Meselson and Franklin Stahl designed an experiment to determine which of these hypotheses would be supported. Nitrogen is a major constituent of DNA. 14N is by far the most abundant isotope of nitrogen, but DNA with the heavier (but non-radioactive) 15N isotope is also functional. Meselson and Stahl grew E. coli in ‘heavy’ nitrogen (15N). What this did was create and E. coli strand with only 15N within the DNA. After growing E. coli in 15N for many generations, the heavy nitrogen E. coli was grown in a normal nitrogen medium 14N. Then, they followed the E. coli for a few generations and could determine the ratio of 15N and 14N in each generation. Comparing this ratio would allow researchers to determine which of these alternate hypotheses would be supported. Let’s see how that would happen.
So when the new generation of E. coli came of age, half of the DNA would parental and half of the DNA would be a copy. Meselson and Wahl could measure the amount of 15N and 14N and determine the percentage of parental vs. daughter chromosomes.This would be true for all of the alternate hypotheses for DNA replication. However, after 2 generations these ratios would be different. Each of these three models makes a different prediction about the distribution of the "15N" DNA in molecules formed after replication. In the conservative hypothesis, after replication, one molecule is the entirely conserved "15N" molecule, and the other is all newly synthesized DNA. The semiconservative hypothesis predicts that each molecule after replication will contain one old and one new strand. This would mean that ½ of the second generation would be low density DNA (those made entirely of 14N), and half would have an intermediate density of DNA. An intermediate density would have ½ of 14N DNA and ½ 15N DNA.
The conservative hypothesis predicts that each double strand of DNA makes an exact copy of itself each generation. If DNA were copied in this way, the 15N and 14N ratio would be different. In the second generation, ¼ of the DNA would be high-density (that is, DNA would be made of 100% 15N DNA), while ¾ of the sample would be low-density DNA (that is, made of 14N DNA). The dispersive model predicts that each strand of each new molecule will contain a mixture of old and new DNA. If DNA were copied via dispersive replication, each generation would consist of intermediate-density DNA. That is all the DNA would be made of parts of the 15N DNA and parts of the 14N DNA.
Results
E. coli were grown for several generations in a medium with 15N. When DNA is extracted from these cells and centrifuged on a salt density gradient, the DNA separates out at the point at which its density equals that of the salt solution. The DNA of the cells grown in15N medium had a higher density than cells grown in normal 14N medium. After that, E. coli cells with only 15N in their DNA were transferred to a 14N medium and were allowed to divide. DNA was extracted periodically and was compared to pure 14N DNA and 15N DNA. After one replication, the DNA was found to have close to the intermediate density. Since conservative replication would result in equal amounts of DNA of the higher and lower densities (but no DNA of an intermediate density), conservative replication was excluded.
However, this result was consistent with both semiconservative and dispersive replication. Semiconservative replication would result in double-stranded DNA with one strand of 15N DNA, and one of 14N DNA, while dispersive replication would result in double-stranded DNA with both strands having mixtures of 15N and 14N DNA, either of which would have appeared as DNA of an intermediate density. The authors continued to sample cells as replication continued. DNA from cells after two replications had been completed was found to consist of equal amounts of DNA with two different densities, one corresponding to the intermediate density of DNA of cells grown for only one division in 14N medium, the other corresponding to DNA from cells grown exclusively in 14N medium. This was inconsistent with dispersive replication, which would have resulted in a single density, lower than the intermediate density of the one-generation cells, but still higher than cells grown only in 14N DNA medium, as the original 15N DNA would have been split evenly among all DNA strands. The result was consistent with the semiconservative replication hypothesis.
DNA polymerase
Meselson and Stahl showed that each parental DNA strand is copied in its entirety; suggesting DNA is replicated via the semiconservative hypothesis. However, they didn’t give a mechanism. The discovery of DNA polymerase cleared the way for the mechanism of DNA synthesis. The discovery of the enzyme, DNA polymerase, led the way to understand more precisely the mechanism by which DNA is replicated. It was discovered that nucleotides are only added to the 3’ end of the nucleic backbone. In this way, DNA is always synthesized from the 5’ to 3’ direction. In other words, DNA is read like a book from front to back (never from back to front).
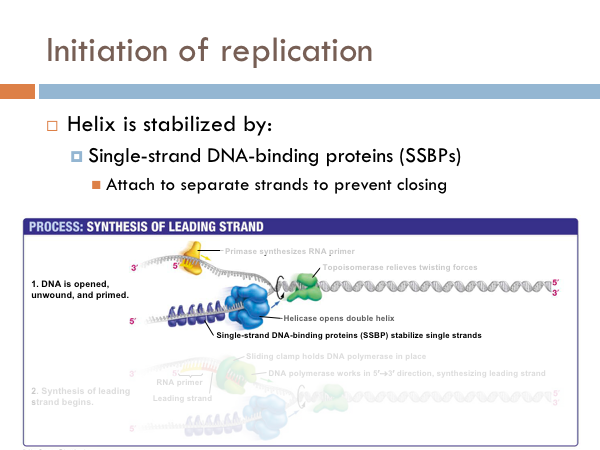
Initiation of replication
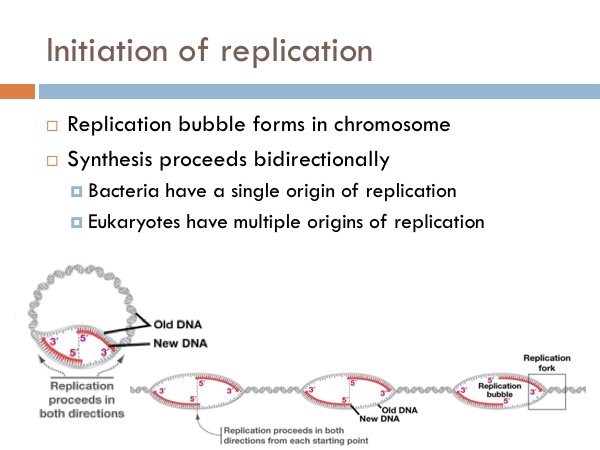
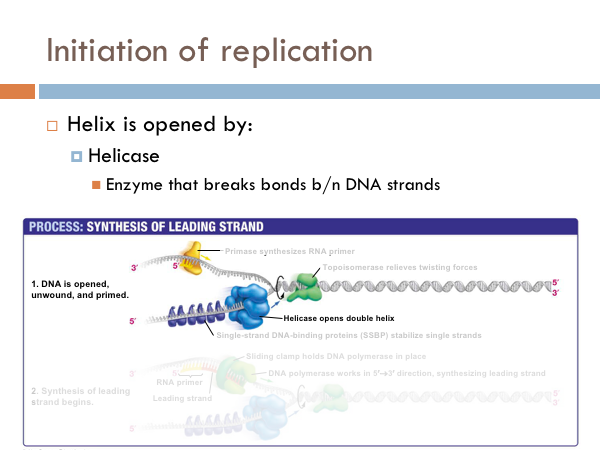
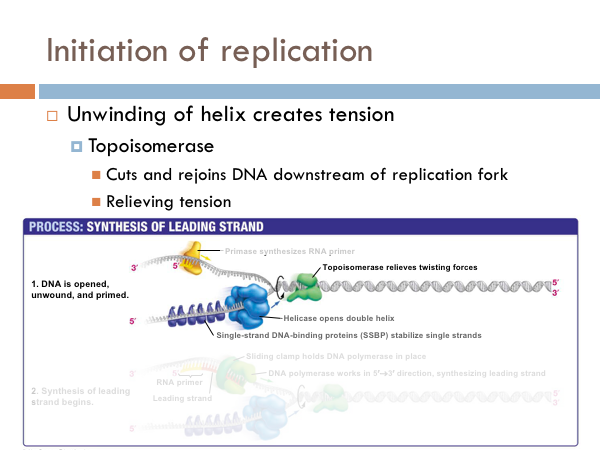
DNA is replicated in replication bubbles in the chromosomes. And synthesis proceeds in two directions. Since replication occurs on from the 5’ to 3’ direction and since DNA is antiparallel, synthesis occurs in both directions. Bacteria have a single origin of replication; in eukaryotes, DNA is replicated at several origins of replication. Let’s look at the details of how DNA is replicated. DNA begins replication by being opened, unwound and primed for replication. The DNA double helix is opened by an enzyme known as helicase. This is a specialized enzyme that breaks the bonds between the two strands of DNA. The DNA helix is then stabilized. This means that special proteins called single-strand DNA binding proteins, attach to the opened DNA strands to prevent them from automatically rebonding. Imagine trying to unwind a slinky into a string of metal with nothing but your hands. You would quickly notice that the unwinding of the slinky would create serious tension. You might employ some tools to help make the process of unwinding the slinky.
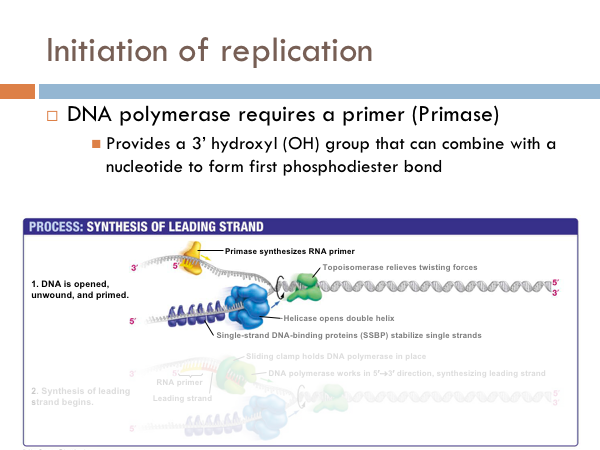
DNA uses a special protein to combat this problem. This protein is called topoisomerase, and it cuts and rejoins the DNA downstream of the replication fork, in effect relieving tension caused by the unwinding of the helix. In the last step of initiation, the DNA polymerase is primed. A special protein, known as primase, starts the process of replication by providing a 3’ hydroxyl group that can a nucleotide in order to form the first phosphodiester bond. Now DNA is ready to be replicated.
Replisome
The replisome is a complex molecular machine that carries out replication of DNA. It is composed of two DNA polymerase complexes, one of which synthesizes the leading strand, while the other synthesizes the lagging strand.
Each dividing nucleoid requires two replisomes for bidirectional replication. The two replisomes continue replication at both forks in the middle of the cell. Finally, as the termination site replicates, the two replisomes separate from the DNA.
The leading strand template is the strand of DNA being replicated continuously. It is the strand that is being continuously polymerized toward the replication fork. All DNA synthesis occurs 5'-3'. The lagging strand grows in the direction opposite to the movement of the growing fork. It grows away from the replication fork and it is synthesized discontinuously. It is called the lagging strand because it synthesized away from the strand, and lags behind the fork.
Synthesis of lagging strand
Because the strand is growing away from the replication fork, it must be replicated in fragments because the Primase (that adds the RNA primer) has to wait until the fork opens to be able to add the primer. Once the Primase attaches the DNA polymerase begins to add bases to the 3’ end of the primer and adds nucleotides away from the replication fork, producing segments of DNA.
These fragments of DNA produced on the lagging strand are called Okazaki fragments. The orientation of the original DNA on the lagging strand prevents continual synthesis. As a result, replication of the lagging strand is more complicated than replication of the leading strand. This process repeats at the fork with further unwinding of the DNA. What is left, is a bunch of Okazaki fragments along the DNA template. Finally, the DNA segments are attached to each other with DNA ligase.
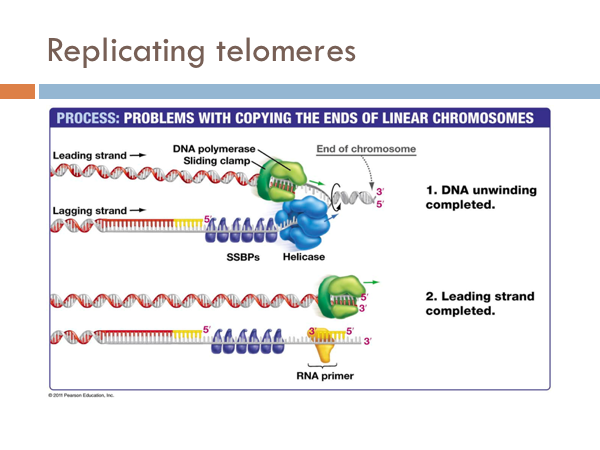
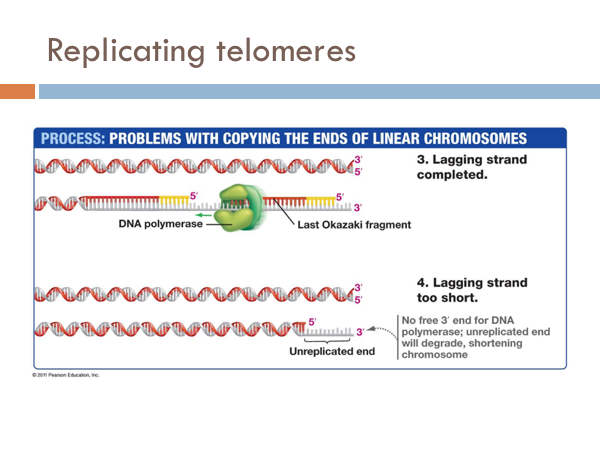
Replicating telomeres
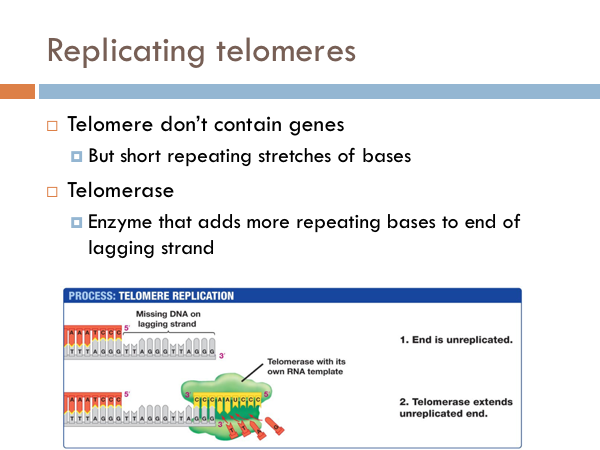
We have seen how DNA is opened, unwound, and primed and how it is replicated on both the leading strand and the lagging strand. Now we find how this process comes to end. The end of the linear chromosomes in eukaryotic organisms are known as telomeres. And at the end, there is no way to replace the RNA primer from the lagging strand because there is no available primer. So the whole primer is removed and leaves a single-strand section of DNA. This is eventually clipped off, which is eventually clipped off, shortening the chromosome. Let’s see what that would look like. At the end of the telomere, the DNA polymerase continues along, business as usual, and slides off completing the leading strand. It is like a zipper without a stopper, it just zips off. Once the last Okazaki fragment is copied, however, it can’t just unzip like the DNA polymerase of the leading strand. This leaves an unreplicated end. This will eventually get cut off. Interestingly, telomeres don’t contain any genes. Rather, they are short repeating stretches of nucleotides. In order that the DNA molecule doesn’t get smaller and smaller and eventually lose all functionality, these repeating nucleotides are copied onto the end of the lagging strand by a special enzyme known as telomerase. This prevents the lagging strand from getting shorter.
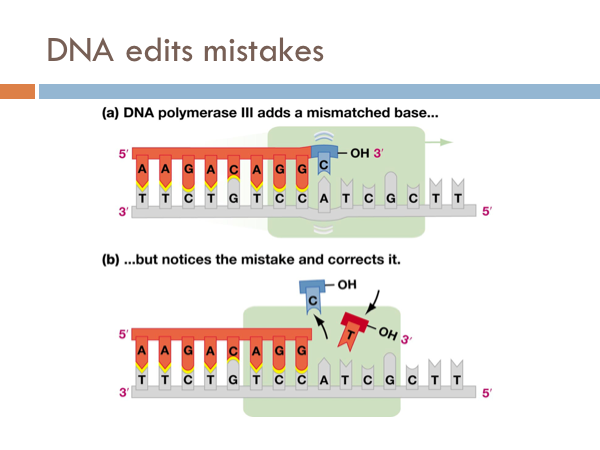
DNA edits mistakes
DNA is incredibly accurate. For every billion bases that are copied only a single base pair is copied wrong. However, even though DNA is this accurate it has a built in spell check. Special enzymes, known as repair enzymes zips down the DNA and removes defective bases and repairs them with their appropriate base pair. This molecule is known as DNA polymerase 3. Even though DNA is extremely efficient at copying itself and it has an built in spell check with DNA polymerase 3, it still makes mistakes. These are known as mutations. Most are inconsequential. Some are detrimental. But every so often, a mutation comes around that actually provides a evolutionary advantage.